The first part of this section describes a concrete proposal
for reproduction. Versions of this have been implemented as prototypes. The
second part discusses possible approaches to the design of other parts of the
language.
4.1 Reproduction
Reproduction is not simply replicating. A reproducing agent
must create an embryo with an appropriate genome and provide resources for
the embryo to grow into an adult agent.
Reproduction has the following steps:
1. Two agents encounter one another (§3.6.5).
2. The agents agree to reproduce and decide who will
be the father (F) and who will be the mother (M).
This is rare in nature (e.g., snails). Better to have a fixed
sex for each agent or allow sex to evolve - but this could be difficult [Taras].
3. F passes its genome to M.
4. M creates a new genome G by combining
the two genomes, using crossover, mutation, etc.
5. M uses G as a program to create a new
agent, the child (C). This may take some time and may consume some of
M's resources.
6. When C is sufficiently grown-up to survive
by itself, M releases it into the environment.
Self-Replication This section is rather technical. It
demonstrates theoretical possibilities rather than something that we might
actually do.
This section is based on early work by von Neumann. We use
strings and functions acting on strings, where we assume that the functions
themselves can be encoded as strings. Upper case letters denote certain fixed
strings and lower case letters represent string variables.
. Infix "+" denotes string concatenation.
. C is a function that copies strings: C(s)
= s.
. B is a function that builds a function (that is,
decodes a string) from its encoded form. It has the property that, for all
strings s and t:
B(s + t) = B(s) +
B(t)
. E is a function that encodes a function as a string.
Thus both B and E are string-to-string functions and E =
B-1.
. K is a control function (actually a combinator)
defined by
K + f + g + x = f(x)
+ g(x)
where f and g are functions and x is a
string.
We consider the effect of K with arguments B,
C, and some string x. We have
K + B + C + x = B(x)
+ C(x). (1)
Let E(K) = k, E(B) = b,
E(C) = c, and x = k + b + c.
Then the right side of (1) simplifies:
B(x) + C(x) = B(k
+ b + c) + x = K + B + C +
x.
Consequently, K + B + C + x
reproduces itself when K is activated.
In order to make this work, we must define a set of
operations, represented as strings, that enable K, B, and C
to be coded with the effects defined above.
If we interpret x = k +b+c as the
"genome" and assume that its position in the agent (that is, its position in the
string K + B + C + x) is known, we can define an
operation activate that applies a string to the genome. To make the agent
reproduce itself, we execute activate K. In turn K executes
activate B and activate C and concatenates the results.
The importance of a mechanism of this kind is that the
agent's ability to reproduce itself is built into it rather than being provided
by the system. This allows the effects of evolution, such as crossover and
mutation, to affect the reproductive process itself.
As an example, we introduce the string "fbgsfgcfcgsfgc.bmbtn.cmotn.gkbc."
which matches the pattern above. (The match is not exact, because the genome has
a prefix "g" and a suffix ".".) The values of K, B, C, and
x are:
K = B(k) = "fbgsfgcfcgsfgc.",
B = B(b) = "bmbtn.",
C = B(c) = "cmotn.",
x = "gkbc.".
This agent replicates itself when its atoms are interpreted
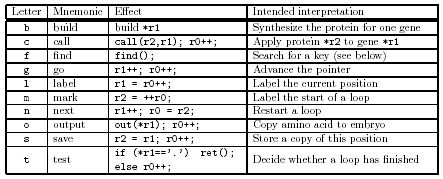
as instructions for a processor. Figure 5 shows both the formal meaning and the
intended interpretation of each atom. The third column shows the effect of the
atom as either pseudocode or C++. Note that some atoms are used as both data and
instructions. Figure 6 explains the reproduction of the agent "fbgsfgcfcgsfgc.bmbtn.cmotn.gkbc.".
The effect of f x is to look for an occurrence of ".
x" in the agent. Each component of the agent starts with "." and a key
letter: for, example, the build starts with "b", the copier starts with "c", and
the genome starts with "g". The sequence "fb" looks for ".b" and starts a new
process at that position of the agent.
In molecular biology, many reactions start because one
molecule binds to another molecule (typically, the molecules are proteins). The
search process performed by "f" is loosely analogous to binding.
Figure 5 gives the appearance that the model is close to
conventional computing, but the following issues should be considered.